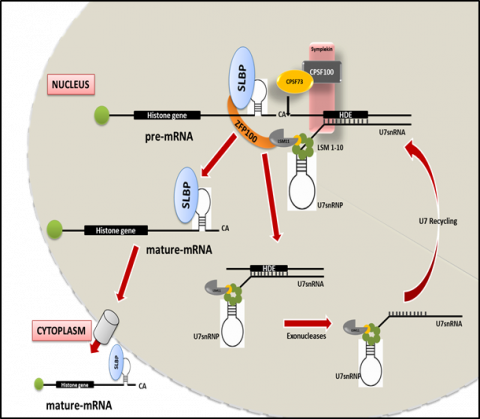
The chromatin state is known to be influenced by multiple mechanisms, including changes in the histone PTMs, incorporation of histone variant and chromatin remodeling, which in turn contributes in governing gene expression patterns and defining cellular identities. Histone isoforms (Canonical) and Histone variants (Non- canonical) can be distinguished by the presence of a conserved stem loop structure at the 3’UTR region of histone isoforms irrespective of the presence of a poly A signal downstream. The stem loop is specifically recognized by a unique protein known as Stem Loop Binding Protein (SLBP). SLBP acts as the key regulator during the three stages of histone mRNA metabolism: pre-mRNA processing, translation and degradation of histone transcripts. Any defect in these processes may lead to an aberrant histone mRNA processing, which might result in a polyadenylated form of histone isoforms, resulting in an improper ratio of histone protein to DNA. Further, this imbalance is known to induce whole genome doublings which induces disease progression.