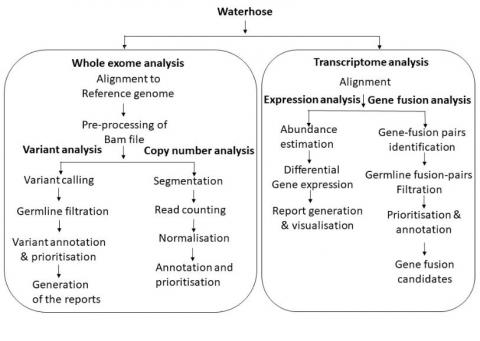
We have developed a fully automated, robust computational pipeline called Waterhose, for analysis of high throughput data generated from paired or orphan tumors. Waterhose provides parallel execution of commands which reduces computation time. It implements filtration of low coverage variants followed by recurrence calculation and annotation of somatic variants using Oncotator and also helps in retrieving already reported and novel somatic variants associated with cancer genome. An additional feature of the waterhose is filtration of genomic alteration specific to the Indian population using the TMC-SNPdb along with dbSNP and ExAc database. To delineate passenger variations from driver events, Waterhose statistically prioritises the driver somatic alteration based on functional predictions algorithms. Furthermore, we have incorporated a separate module that helps in copy number analysis which makes it more comprehensive for whole- genome or exome analysis. In summary, Waterhose is a simple graphical user interface based fully automated framework that generates graphical output in form of heat map which makes it more user friendly for non-computational biologist to analyse cancer genomes in more efficient manner.
Figure: Overall schema of Waterhose